Non-Uniform Sampling (NUS) is available for CP800 spectrometer
(NUS Data processing can be done on either CP800 computer or BioNMR PC in the NMR room)
Non-uniform sampling (NUS) is a method of acquiring data using less time than the conventional NMR methods. The method can also be used to improve resolution if experiment time is limited. The following has two brief guides of NUS paramenter set up, hmsIST and TopSpin.
nus@HMS Parameter Setup
TopSpin NUS Setup
1. Click AcquPars, and change FnTYPE to non-uniform_sampling.
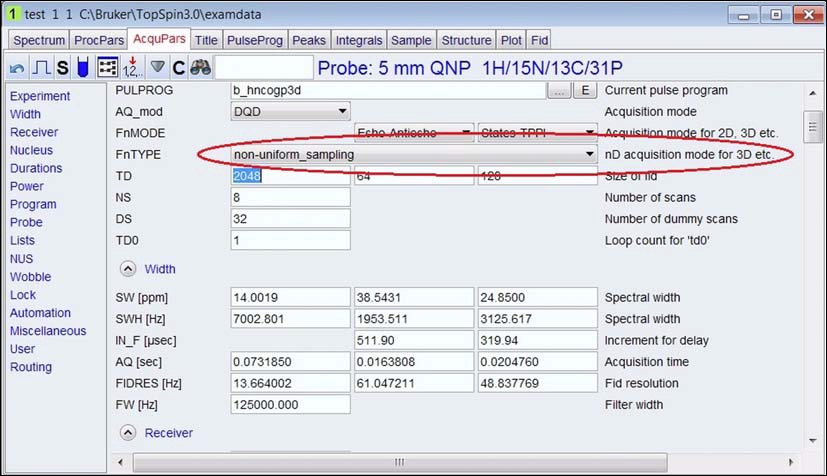
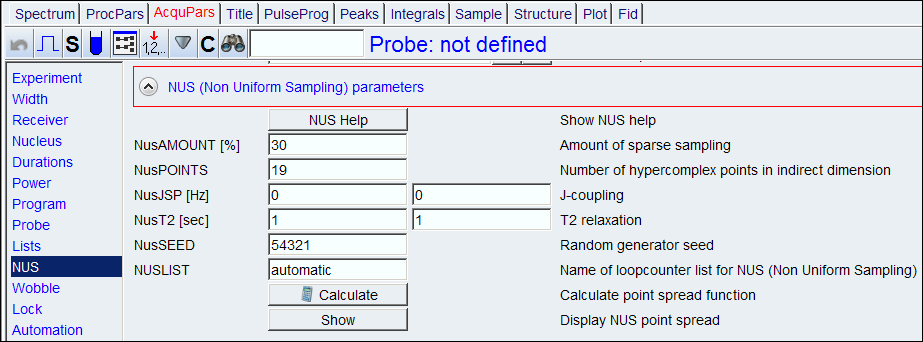
2. Go to NUS section, set NusAMOUNT[%]. The amount is usually 50% or less, and make sure NUSLIST is set automatic.
3. Run the experiment as usual.
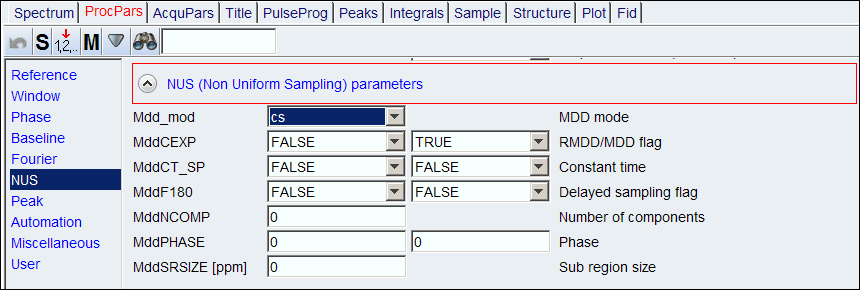
4. To process the spectrum, go to ProcPars page, and at NUS section, set MDD_mod to either cs or mdd (cs may give better result). Then, process the data as usual.
3D NUS Data phasing
(1) Using a third party software to process data if available, such as hmsIST or Mnova
(2) TopSpin method.
(a) It is highly recommended to acquire a 2D plane first in conventional mode to determine the phase correction.
(b) You may read the first FID, "reser 1"; determine the phase for f3 from this 1D spectrum. If the experiment is ET or Echo Anti-Echo, you need to add or subtract 90 degrees from the phc0 value determined from the 1D.
(c) The indirect dimensions should not require a phase correction.
- In TopSpin AcquPars window, change the FnTYPE to be 'non-uniform_sampling'. You don’t need to set anything in the NUS parameters block.
- Execute the macro by typing 'nusPGSv3'. It will ask several questions.
-
- Random Seed
- - 0 is default and probably what you want.
- - 0 implies the random seed used for random number generation will be selected by the computer.
- - Any other number here means you will also get the same sequence. So lets say you want to compare two acquisitions but are not interested in the differences that might result from sampling differently.
-
- Sine portion for sampling
- - 2 is default and probably what you want.
- - This is not the place to describe what this means. 2 will result in more points towards the front of the schedule and this is probably best for you. 1 and 0 are experimental and we don't suggest you use them unless you know why you want to.
-
- Number of points to collect (complex points)
- - When set 128 out of 1280, it is 10% sampling amount. 10% of the current numbers of linear, complex points is selected by default.
- - When running a 2D experiment, you probably want this to be more like 25%. 10% of all the complex points should work for a 3D experiment. 1-2% should work for a 4D. Not that the number you put in here is the total number of points to be sampled, not the percentage.
-
- Tolerance (as a percentage).
- - 0.01 % is the default. This means the generator must generate the number of points requested above +/- 0.01%. Most likely this means it will generate exactly the number you asked. Sometimes it is hard for the scheduler to find the exact number of samples you ask for so you may need to add some tolerance here. Try changing it to 10%.
-
- Shuffle the schedule (0 = no, 1 = yes)
- - Default is 0 which means the schedule will be generated with the left most column being "fast" and the right most column being the slowest. I suggest this for starting out.
- - Select 1 will shuffle the schedule. It is perfectly fine to do this as the reconstruction processing will reorder everything in the end anyway. However, note that the first point, (0 or 0,0 or 0,0,0) will always remain at the top.
- - At the end there should be a message” Poisson gas sampling schedule setup finished.”. A nusPGS_setup.out file will be generated into the data directory.
-
- Collect data with the 'zg' command. The schedule used will be deposited into the data directory with the name 'nuslist'. You can use this schedule as is during reconstruction with hmsIST.
- NUS data processing: http://gwagner.med.harvard.edu/intranet/hmsIST/234Proc.html. nmrPipe, nmrDraw and a C shell installed on your linux or Mac computer is required.