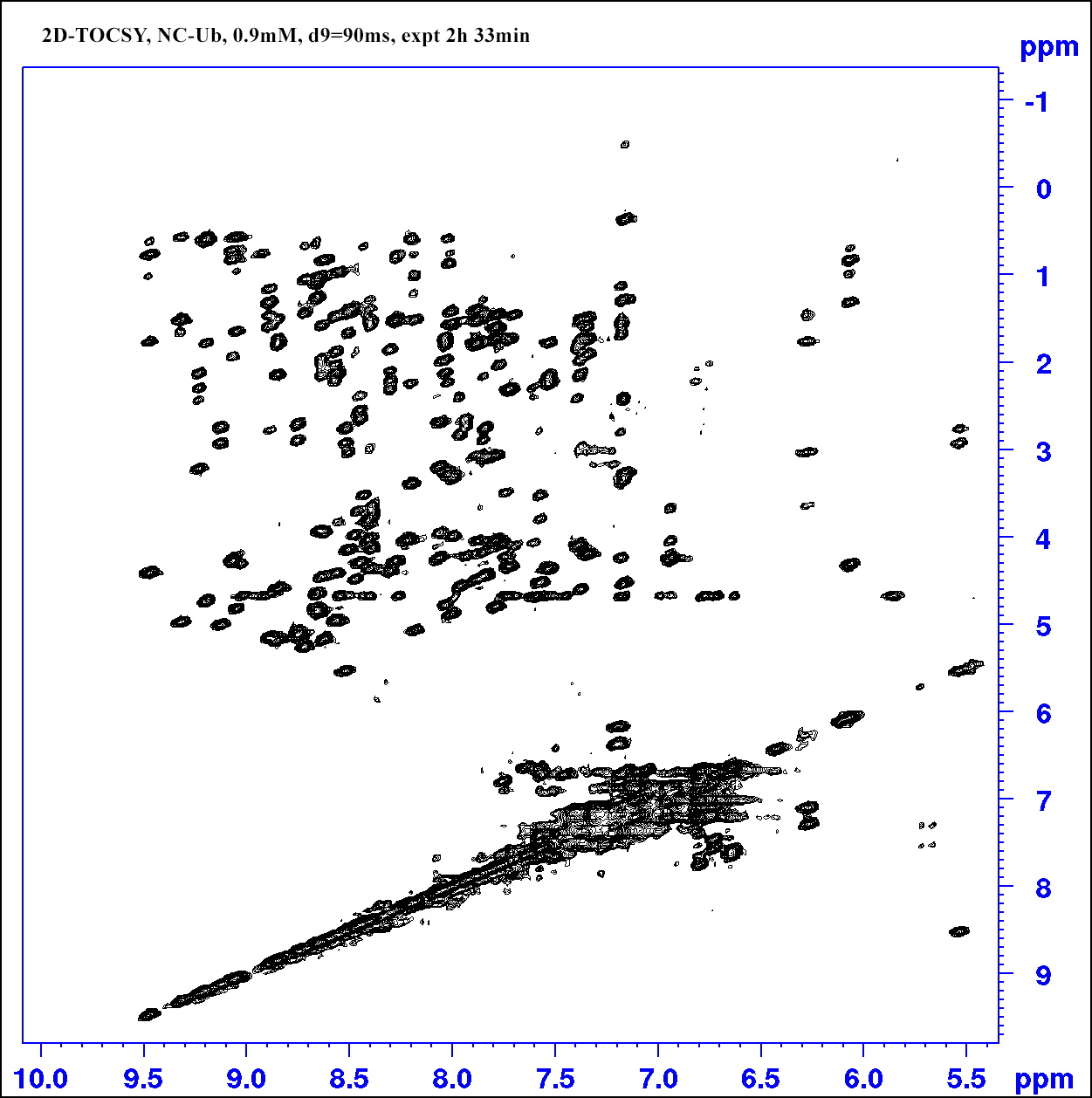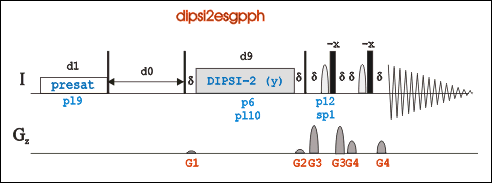
Go to an existing 2D-Tocsy experiment (pulse program: dipsi2esfbgpph), type “edc” to copy its parameters and generate a new experiment number. Or in a new experiment, "rpar DIPSI2ESFBGPPH".
Pulse program dipsi2esfbgpph is based on dipsi2esgpph and have f2 and f3 channels included.
-
Type "edasp" to check /adjust spectrometer routing.
-
Type “eda”, set parameters td, ns, ds, sw, O1P, O2P and O3P if necessary.
-
In acquisition window, if sample is unlabeled, leave ZGOPTNS blank. If sample is N15 labeled, set ZGOPTNS "-DLABEL_N". If sample is N15 and C13 labeled, set ZGOPTNS "-DLABEL_CN".
-
Type “ased”, adjust power length and power level for 1H. If sample is labeled, you will need to check 15N and/or 13C power level and power length according. Or type "getprosol 1H A B" to set power related parameters. A is the pulse length in µs, and B is the power leverl in dB.
-
Type "pulse" to calculate pL10 based on targeting p6 value. Use p6 35 us.
-
Set d9 (Tocsy mixing time) 90 ms for a protein sample; set pL32 70dB for water presaturation
-
In gs mode, adjust O1P and p12 to achieve minimum FID area integral for water suppression.
-
Start the experiment by rga and zg.
-
Process the FID by typing “xfb”. Phase the spectrum when necessary.