
Education
- 1984 – 1987, Dept. of Biochemistry, Stanford University. Research topic: Enzymes of DNA recombination in E. coli. Ph. D., Biochemistry, 1984, University of California, Berkeley. Dissertation topic: Enzyme mechanisms and kinetic isotope effects.
- B.A., Chemistry, 1978, Haverford College, Haverford, PA.
Professional Experience
- Associate Professor, 1993 – present, Department of Chemistry and Biochemistry, University of Maryland.
- Assistant Professor, 1987 – 1993, Department of Chemistry and Biochemistry, University of Maryland. Member, Program in Molecular and Cellular Biology, University of Maryland.
Research Interests
Enzymology and mechanisms of DNA recombination and repair. Mechanism of the RecBCD enzyme from E. coli. DNA repair in Deinococcus radiodurans
Professional Societies
American Society for Biochemistry and Molecular Biology, American Society for Microbiology, American Chemical Society, American Association for the Advancement of Science for Microbiology
Major Recognitions and Honors
- Postdoctoral Fellowship, Damon Runyon-Walter Winchell Cancer Fund, 1984 – 1987
- NSF Graduate Fellowship, 1978 – 1981
- Phi Beta Kappa, 1977
Significant Professional Service and Activities
Workshop chair, “Enzymology of Homologous Recombination,” Keystone Symposium on Molecular Mechanisms of DNA Replication and Recombination, 2002. Grant review panel, US DOE, Experimental and Computational Structural Biology, 2000. Biochemistry Study Section, NIH, June, 2002; June, 2003; Oct. 2004. Microbial Physiology and Genetics Study Section (MCB-2), NIH, Oct., 2003. Minority Biomedical Research Support Review Panel, NIH, June, 2005. Molecular Genetics (A) Study Section, NIH, Feb., 2006. Board of Scientific Counselors, NIDDK, NIH (Ad hoc member), April, 2007. Consultant, Biohelix Corp., Beverly, MA. Review manuscripts for Cell, Biochemistry, J. Biol. Chem., J. Bacteriol., Genes & Devel., Proc. Natl. Acad. Sci., USA, Molec. Microbiol., Genetics, BMC Microbiology, etc.
Students Mentored
Dr. Julin has mentored 14 students who received a PhD in Biochemistry or in Molecular and Cellular Biology, and 5 students who received MS degrees in Biochemistry.
Living organisms are under constant assault by environmental agents that damage their DNA, including radiation and reactive chemicals. These agents lead to formation of damaged bases and DNA strand breaks which, if unrepaired, can lead to cell death, mutation, and cancer in humans. Organisms defend themselves against this onslaught by deploying a variety of DNA repair and recombination enzymes that are able to undo the damage and restore the DNA to its original undamaged state. We are studying enzymes that participate in DNA repair in the bacterium Deinococcus radiodurans, and the RecBCD enzyme from Escherichia coli.
DNA Repair in Deinococcus radiodurans
D. radiodurans is a fascinating bacterium because of its ability to survive under extreme conditions that are lethal to most other forms of life. The organism, dubbed “Conan the Bacterium”, can withstand high levels of ionizing radiation (X-rays and gamma rays), ultraviolet light, and DNA-damaging chemicals. The physiological properties and enzymatic pathways that enable it to survive these conditions are of great interest.
The most serious consequence of ionizing radiation is the formation of double-strand breaks in DNA. D. radiodurans can survive a much greater number of breaks in its chromosomes than can E. coli and other organisms, suggesting that it has very efficient pathways for repair of these breaks. We are interested in identifying the enzymes that enable D. radiodurans to repair massive numbers of double-strand breaks to its chromosomes. A surprising fact about D. radiodurans is that it lacks a homologue of the RecBCD enzyme which is essential for ds-break repair in other bacteria (see below). RecBCD is both a helicase and a nuclease that acts at double-strand DNA breaks in the initial steps of double-strand break repair. We are particularly interested in identifying and studying the helicase(s) and nuclease(s) that may substitute for RecBCD in D. radiodurans. Our studies show that several enzymes, including the RecD helicase, a distant homologue of helicase IV, and the RecJ exonuclease, contribute to double-strand break repair in the absence of RecBCD. We use both in vivo and in vitro approaches to study these enzymes. Deletion mutants can be generated using homologous recombination techniques. The phenotypes of the mutants can then be studied using a variety of assays that test sensitivity of the cells to DNA damaging agents. We also express and purify the enzymes and study their biochemical and enzymatic properties.
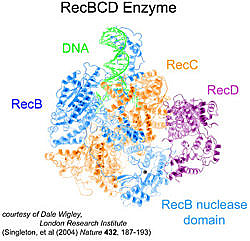 E. coli RecBCD enzyme
E. coli RecBCD enzyme
RecBCD is a complex macromolecular machine composed of three protein subunits called the RecB, RecC, and RecD proteins. The enzyme is essential for the repair of double-strand breaks in E. coli and many other bacteria. RecBCD binds to the end of a broken DNA duplex and acts as an ATP-dependent helicase and nuclease to process the DNA for subsequent repair via homologous recombination.
We discovered in previous work that the nuclease activity is localized to a small domain that is part of the RecB subunit. The complete structure of this domain was revealed in the crystal structure. Our current research focuses mainly on the properties of the RecB nuclease domain and how it interacts with the other activities of RecBCD during the complex reaction with double-stranded DNA. We use enzyme kinetics, site-directed muta-genesis, protein chemistry, and other methods.


